Materials+ML Workshop Day 5¶
- The Atomic Simulation Environment
- Building atomic structures
- Visualizing atomic structures
- Python Materials Genomics (
pymatgenwithase)- Visualizing properties (band structure)
- Using the Materials Project Database
- Querying material properties
- Getting crystal structure data
The Workshop Online Book:¶
https://cburdine.github.io/materials-ml-workshop/¶
Tentative Week 1 Schedule:¶
| Session | Date | Content |
| Day 1 | 06/09/2025 (2:00-4:00 PM) | Introduction, Python Data Types |
| Day 2 | 06/10/2025 (2:00-4:00 PM) | Python Functions and Classes |
| Day 3 | 06/11/2025 (2:00-4:00 PM) | Scientific Computing with Numpy and Scipy |
| Day 4 | 06/12/2025 (2:00-4:00 PM) | Data Manipulation and Visualization |
| Day 5 | 06/13/2025 (2:00-4:00 PM) | Materials Science Packages, Introduction to ML |
Review: Day 4¶
Pandas¶
- Pandas is an open-source Python package for data manipulation and analysis.
- It can be used for reading writing data to several different formats including:
- CSV (comma-separated values)
- Excel spreadsheets
- SQL databases
- We can import pandas as follows:
In [1]:
import pandas as pd
DataFrames¶
- We can create Dataframes from Python dictionaries as follows:
In [2]:
# Data on the first four elements of the periodic table:
elements_data = {
'Element' : ['H', 'He', 'Li', 'Be'],
'Atomic Number' : [ 1, 2, 3, 4 ],
'Mass' : [ 1.008, 4.002, 6.940, 9.012],
'Electronegativity' : [ 2.20, 0.0, 0.98, 1.57 ]
}
# construct dataframe from data dictionary:
df = pd.DataFrame(elements_data)
In [6]:
import numpy as np
# get the 'Mass' column and convert it to a numpy array:
mass_series = df['Mass']
mass_array = np.array(mass_series)
print(mass_array)
[1.008 4.002 6.94 9.012]
In [28]:
# function for estimating mass:
def amu_mass(n):
return 2*n
# add an "Estimated Mass" column to the dataframe:
df['Estimated Mass'] = \
df['Atomic Number'].apply(amu_mass)
display(df)
| Element | Atomic Number | Mass | Electronegativity | Estimated Mass | |
|---|---|---|---|---|---|
| 0 | H | 1 | 1.008 | 2.20 | 2 |
| 1 | He | 2 | 4.002 | 0.00 | 4 |
| 2 | Li | 3 | 6.940 | 0.98 | 6 |
| 3 | Be | 4 | 9.012 | 1.57 | 8 |
Matplotlib¶
- Matplotlib is a MATLAB-like plotting utility for creating publication-quality plots
- In matplotlib, we typically import the
pyplotsubpackage with the aliasplt:
In [8]:
import matplotlib.pyplot as plt
In [10]:
# generate some data:
data_x = np.linspace(0,8,10)
data_y = np.sin(data_x)
# create a new figure and plot data:
plt.figure(figsize=(7,2))
plt.grid()
plt.plot(data_x, data_y, 'ro--')
# add a title and show plot in notebook:
plt.title('Example of a Line plot')
plt.show()
New Content:¶
- Materials Science Python Packages:
- ASE (Atomic Simulation Environment)
- Pymatgen (Python Materials Genomics)
- Materials Project API
Installing packages:¶
- Install ASE:
pip install ase
- Install ASE:
pip install pymatgen
- Install Materials Project API
pip install mp-api
In [25]:
pip install ase pymatgen mp-api
Defaulting to user installation because normal site-packages is not writeable Requirement already satisfied: mp-api in /home/colin/.local/lib/python3.10/site-packages (0.33.3) Requirement already satisfied: msgpack in /usr/lib/python3/dist-packages (from mp-api) (1.0.3) Requirement already satisfied: typing-extensions>=3.7.4.1 in /home/colin/.local/lib/python3.10/site-packages (from mp-api) (4.2.0) Requirement already satisfied: pymatgen>=2022.3.7 in /home/colin/.local/lib/python3.10/site-packages (from mp-api) (2023.5.31) Requirement already satisfied: monty>=2021.3.12 in /home/colin/.local/lib/python3.10/site-packages (from mp-api) (2023.5.8) Requirement already satisfied: emmet-core>=0.54.0 in /home/colin/.local/lib/python3.10/site-packages (from mp-api) (0.57.1) Requirement already satisfied: setuptools in /usr/lib/python3/dist-packages (from mp-api) (59.6.0) Requirement already satisfied: requests>=2.23.0 in /usr/lib/python3/dist-packages (from mp-api) (2.25.1) Requirement already satisfied: spglib>=2.0.1 in /home/colin/.local/lib/python3.10/site-packages (from emmet-core>=0.54.0->mp-api) (2.0.2) Requirement already satisfied: pybtex~=0.24 in /home/colin/.local/lib/python3.10/site-packages (from emmet-core>=0.54.0->mp-api) (0.24.0) Requirement already satisfied: pydantic>=1.10.2 in /home/colin/.local/lib/python3.10/site-packages (from emmet-core>=0.54.0->mp-api) (1.10.9) Requirement already satisfied: ruamel.yaml>=0.17.0 in /home/colin/.local/lib/python3.10/site-packages (from pymatgen>=2022.3.7->mp-api) (0.17.32) Requirement already satisfied: networkx>=2.2 in /home/colin/.local/lib/python3.10/site-packages (from pymatgen>=2022.3.7->mp-api) (2.8.8) Requirement already satisfied: palettable>=3.1.1 in /home/colin/.local/lib/python3.10/site-packages (from pymatgen>=2022.3.7->mp-api) (3.3.3) Requirement already satisfied: scipy>=1.5.0 in /usr/lib/python3/dist-packages (from pymatgen>=2022.3.7->mp-api) (1.8.0) Requirement already satisfied: sympy in /usr/lib/python3/dist-packages (from pymatgen>=2022.3.7->mp-api) (1.9) Requirement already satisfied: tqdm in /home/colin/.local/lib/python3.10/site-packages (from pymatgen>=2022.3.7->mp-api) (4.64.0) Requirement already satisfied: numpy>=1.20.1 in /usr/lib/python3/dist-packages (from pymatgen>=2022.3.7->mp-api) (1.21.5) Requirement already satisfied: plotly>=4.5.0 in /home/colin/.local/lib/python3.10/site-packages (from pymatgen>=2022.3.7->mp-api) (5.15.0) Requirement already satisfied: uncertainties>=3.1.4 in /home/colin/.local/lib/python3.10/site-packages (from pymatgen>=2022.3.7->mp-api) (3.1.7) Requirement already satisfied: pandas in /home/colin/.local/lib/python3.10/site-packages (from pymatgen>=2022.3.7->mp-api) (1.4.4) Requirement already satisfied: tabulate in /home/colin/.local/lib/python3.10/site-packages (from pymatgen>=2022.3.7->mp-api) (0.9.0) Requirement already satisfied: matplotlib>=1.5 in /usr/lib/python3/dist-packages (from pymatgen>=2022.3.7->mp-api) (3.5.1) Requirement already satisfied: packaging in /usr/lib/python3/dist-packages (from plotly>=4.5.0->pymatgen>=2022.3.7->mp-api) (21.3) Requirement already satisfied: tenacity>=6.2.0 in /home/colin/.local/lib/python3.10/site-packages (from plotly>=4.5.0->pymatgen>=2022.3.7->mp-api) (8.2.2) Requirement already satisfied: PyYAML>=3.01 in /usr/lib/python3/dist-packages (from pybtex~=0.24->emmet-core>=0.54.0->mp-api) (5.4.1) Requirement already satisfied: six in /usr/lib/python3/dist-packages (from pybtex~=0.24->emmet-core>=0.54.0->mp-api) (1.16.0) Requirement already satisfied: latexcodec>=1.0.4 in /home/colin/.local/lib/python3.10/site-packages (from pybtex~=0.24->emmet-core>=0.54.0->mp-api) (2.0.1) Requirement already satisfied: ruamel.yaml.clib>=0.2.7 in /home/colin/.local/lib/python3.10/site-packages (from ruamel.yaml>=0.17.0->pymatgen>=2022.3.7->mp-api) (0.2.7) Requirement already satisfied: future in /home/colin/.local/lib/python3.10/site-packages (from uncertainties>=3.1.4->pymatgen>=2022.3.7->mp-api) (0.18.3) Requirement already satisfied: python-dateutil>=2.8.1 in /home/colin/.local/lib/python3.10/site-packages (from pandas->pymatgen>=2022.3.7->mp-api) (2.8.2) Requirement already satisfied: pytz>=2020.1 in /usr/lib/python3/dist-packages (from pandas->pymatgen>=2022.3.7->mp-api) (2022.1) Note: you may need to restart the kernel to use updated packages.
The Atomic Simulation Environment¶
- ASE is a Python package for building, manipulating, and performing calculations on atomic structures
- ASE provides interfaces to several different simulation platforms, such as:
- VASP
- Quantum ESPRESSO
- Q-Chem
- Gaussian
ASE Basics:¶
The fundamental data type in ASE is the
Atomsobject:Atomsrepresents a collection of Atoms in a molecular or crystalline structure.- Material properties (such as the results of calculations) can be attached to Atoms instances
- ASE has functionality for loading, exporting and viewing
Atomsobjects in different formats.
In [25]:
from ase.build import molecule
from ase.visualize import view
# build a common molecule:
acetic_acid = molecule('CH3COOH')
view(acetic_acid, viewer='x3d')
Out[25]:
Tutorial: ASE - The Atomic Simulation Environment¶
- Building simple molecules
- Building inorganic structures
- MXenes
- Carbon Nanotubes
Exercise: ASE - The Atomic Simulation Environment¶
- Nitrogen-Vacancy Centers in Diamond
The Materials Project¶
- Register for an account at https://next-gen.materialsproject.org/
- Once you have registered, find your API key under My Dashboard:
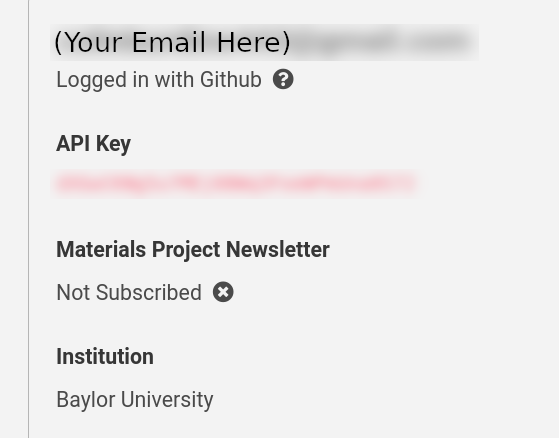
- Once you have copied your API key, paste it into your notebook as a string:
In [30]:
# create a variable in your notebook with your API key
MP_API_KEY = '< Paste your API key here. >'
Always avoid sharing your API key with others.
Tutorial: Working with the Materials Project¶
- We will examine the material
YBa$_1$Cu$_2$O$_7$ (YBCO)
- YBCO is a high-temperature superconductor
- We will:
- Visualize the crystal structure
- Plot the Band Structure
Exercise: Working with the Materials Project¶
- Visualize the DOS of YBa$_2$Cu$_3$O$_7$
Questions¶
- Any questions about Week 1 or Week 2 Content?
Recommended Reading:¶
- Introduction to Machine Learning
- Statistics Review (as needed)
- Mathematics Review (as needed)
- Supervised Learning
If possible, try to do the exercises. Bring your questions to our next meeting (next Monday).